📄 Domain and Content Adaptive Convolution based Multi-Source Domain Generalization for Medical Image Segmentation (DCAC)
This repo contains the official implementation of our paper: Domain and Content Adaptive Convolution based Multi-Source Domain Generalization for Medical Image Segmentation, which adapts dynamic convolution for domain generalization.
(1) Multi-scale features based domain relationship modeling.
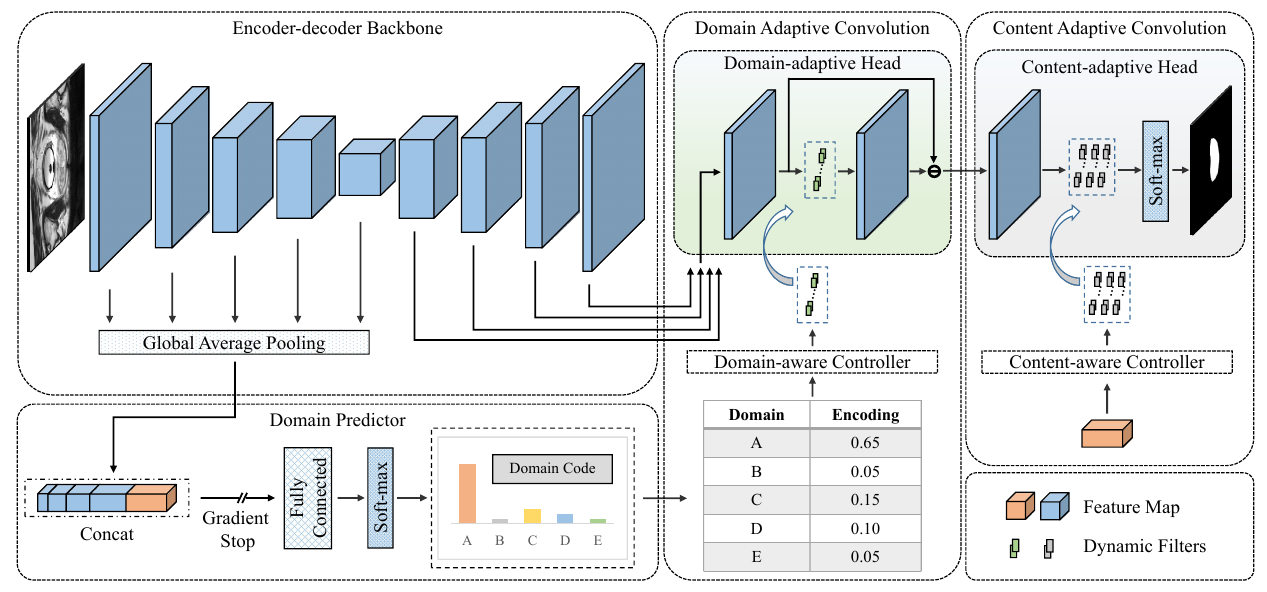
We use the domain-discriminative information embedded in the encoder feature maps to generate the domain code of each input image, which establishes the relationship between multiple source domains and the unseen target domain.
(2) Domain and Content Adaptive Convolution.
We design the dynamic convolution-based domain adaptive convolution (DAC) module and content adaptive convolution (CAC) module to enable our DCAC model to adapt not only to the unseen target domain but also to each test image.
(3) Competitive results on three benchmarks.
We present extensive experiments, which demonstrate the effectiveness of our DCAC model against the state-of-the-art in three medical image segmentation benchmarks with different imaging modalities.
This repo was tested with Ubuntu 20.04.3 LTS, Python 3.8, PyTorch 1.8.0, and CUDA 10.1. But it should be runnable with Ubuntu 16.04 and Ubuntu 18.04.
We suggest using virtual env to configure the experimental environment. Compiling PyTorch on your own workstation is suggested but not needed.
- Clone this repo:
git clone https://github.com/ShishuaiHu/DCAC.git- Create experimental environment using virtual env:
cd DCAC/nnUNet
virtualenv .env --python=3.8 # create
source .env/bin/activate # activate
bash ./install.sh # install torch and nnUNet (equipped with DCAC)- Configure the paths in
.envrcto the proper path:
echo -e '
export nnUNet_raw_data_base="nnUNet raw data path you want to store in"
export nnUNet_preprocessed="nnUNet preprocessed data path you want to store in, SSD is prefered"
export RESULTS_FOLDER="nnUNet trained models path you want to store in"' > .envrc
source .envrc # make the variables take effectThe dataset details and the download link can be found in the Project Page.
python nnunet/dataset_conversion/Task1001_Prostate.py
python nnunet/dataset_conversion/Task1007_COVID_19.py
python nnunet/dataset_conversion/Task1011_Fundus.py
# Prostate
nnUNet_plan_and_preprocess -t 1001
nnUNet_plan_and_preprocess -t 1002
nnUNet_plan_and_preprocess -t 1003
nnUNet_plan_and_preprocess -t 1004
nnUNet_plan_and_preprocess -t 1005
nnUNet_plan_and_preprocess -t 1006
# COVID_19
nnUNet_plan_and_preprocess -t 1007
nnUNet_plan_and_preprocess -t 1008
nnUNet_plan_and_preprocess -t 1009
nnUNet_plan_and_preprocess -t 1010
# Fundus
nnUNet_plan_and_preprocess -t 1011
nnUNet_plan_and_preprocess -t 1012
nnUNet_plan_and_preprocess -t 1013
nnUNet_plan_and_preprocess -t 1014# Prostate
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Prostate 1001 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Prostate 1002 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Prostate 1003 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Prostate 1004 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Prostate 1005 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Prostate 1006 all
# COVID_19
CUDA_VISIBLE_DEVICES=0 nnUNet_train 3d_fullres DCACTrainer_COVID 1007 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 3d_fullres DCACTrainer_COVID 1008 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 3d_fullres DCACTrainer_COVID 1009 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 3d_fullres DCACTrainer_COVID 1010 all
# Fundus
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Fundus 1011 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Fundus 1012 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Fundus 1013 all
CUDA_VISIBLE_DEVICES=0 nnUNet_train 2d DCACTrainer_Fundus 1014 all# Prostate
CUDA_VISIBLE_DEVICES=0 nnUNet_predict -i $nnUNet_raw_data_base/nnUNet_raw_data/Task1001_Target_BIDMC/imagesTs -o $RESULTS_FOLDER/nnUNet/2d/Task1001_Target_BIDMC/DCACTrainer_Prostate__nnUNetPlansv2.1/predictions -t 1001 -m 2d -tr DCACTrainer_Prostate --save_npz -f all
# Others are similar to this commandIf you find this repo useful for your research, please consider citing the paper as follows:
@article{hu2022domain,
title={Domain and Content Adaptive Convolution based Multi-Source Domain Generalization for Medical Image Segmentation},
author={Hu, Shishuai and Liao, Zehui and Zhang, Jianpeng and Xia, Yong},
journal={IEEE Transactions on Medical Imaging},
year={2022},
publisher={IEEE}
}